Finite State Projection of a Gene Transmission Model
Model
In this application example, we consider the two-stage model of gene transmission, discussed e.g. in. [1]. The two-stage model describes the stochastic transcription and translation of a gene by the reactions
The solution to the Chemical Master Equation (CME) [2] gives the probabilities of the states at a given time point as the solution of an infinite dimensional ODE
We assume the initial probabilites to be independent Poisson distributions for mRNA and Protein abundances.
Finite State Projection
The Finite State Projection [3] approximates this CME by restricting the states to a finite domain and approximating the probability of the left out states by zero. In our case, we ill restrict the states to \(x_{r, p}\) for \((r, p) \in [0, r_{max}-1] \times [[0, p_{max}-1]]\).
Model Construction:
The model is constructed using the yaml2sbml Model Editor and exported to SBML. This takes only a few lines of code:
[1]:
from yaml2sbml import YamlModel
from itertools import product
from scipy.stats import poisson
model = YamlModel()
r_max = 20
p_max = 50
lambda_r = 5
lambda_p = 5
# add parameters
model.add_parameter(parameter_id='k_1', nominal_value=2)
model.add_parameter(parameter_id='k_2', nominal_value=1)
model.add_parameter(parameter_id='k_3', nominal_value=10)
model.add_parameter(parameter_id='k_4', nominal_value=3)
# add ODEs & construct the rhs
for r, p in product(range(r_max), range(p_max)):
rhs = f'-(k_1 + (k_2 + k_3)*{r} + k_4*{p}) * x_{r}_{p} '
if r>0:
rhs += f'+ k_1 * x_{r-1}_{p}'
if r+1 < r_max:
rhs += f'+ k_2 * {r+1} * x_{r+1}_{p}'
if p>0:
rhs += f'+ k_3 * {r} * x_{r}_{p-1}'
if p+1 < p_max:
rhs += f'+ k_4 * {p+1} * x_{r}_{p+1}'
model.add_ode(state_id = f"x_{r}_{p}",
right_hand_side=rhs,
#convert from np.float to float
initial_value=float(poisson.pmf(r, lambda_r)*poisson.pmf(p, lambda_p))
)
model.write_to_sbml('gene_expression.xml', overwrite=True)
Model Simulation
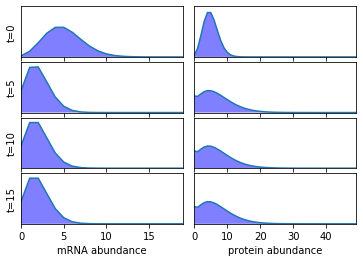
We now use AMICI to simulate the generated model and plot the marginal distributions of the mRNA and protein abundance.
[2]:
%matplotlib inline
import numpy as np
from utils import plot_AMICI
# sim, model = compile_and_simulate('gene_expression.xml', 0.1 * np.arange(10), r_max, p_max)
plot_AMICI('gene_expression.xml', [0, 5, 10, 15], r_max, p_max)
References
[1] Shahrezaei, V., and Swain, P.S. (2008). “Analytical distributions for stochastic gene expression” PNAS 17256–17261 https://www.pnas.org/content/105/45/17256
[2] Gillespie, D. T. (1992). “A rigorous derivation of the chemical master equation” Physica A: Statistical Mechanics and its Applications https://www.sciencedirect.com/science/article/abs/pii/037843719290283V
[3] Munsky, B. and Khammash, M. (2006) “The finite state projection algorithm for the solution of the chemical master equation” The Journal of chemical physics https://aip.scitation.org/doi/full/10.1063/1.2145882